This page provides an introductory quick-start tutorial on running an NBO calculation and interpreting the output. The example chosen is that of methylamine (CH3NH2) in Pople-Gordon idealized geometry, treated at the ab initio RHF/3-21G level. This simple split-valence basis set consists of 28 AOs (nine each on C and N, two on each H), extended by 13 AOs beyond the minimal basis level.

Input files to perform this calculation are given here for Gaussian and GAMESS. The pop=nbo option of the Gaussian program requests default NBO analysis. NBO analysis is requested in the GAMESS calculation by simply including the line
$NBO $END
in the GAMESS input file. This "empty" NBO keylist specifies that NBO analysis should be carried out at the default level.
The default NBO output produced by this example is shown below, just as it appears in your output file. The start of the NBO section is marked by a standard header, citation, job title, and storage info:
*********************************** NBO 5.0 ***********************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
*******************************************************************************
(c) Copyright 1996-2001 Board of Regents of the University of Wisconsin System
on behalf of the Theoretical Chemistry Institute. All Rights Reserved.
Cite this program as:
NBO 5.0. E. D. Glendening, J. K. Badenhoop, A. E. Reed,
J. E. Carpenter, J. A. Bohmann, C. M. Morales, and F. Weinhold
(Theoretical Chemistry Institute, University of Wisconsin,
Madison, WI, 2001)
Job title: Methylamine...RHF/3-21G//Pople-Gordon standard geometry
Storage needed: 2562 in NPA, 3278 in NBO ( 2000000 available)
Note that all NBO output is formatted to a maximum 80-character width for convenient display. The NBO heading echoes any requested keywords (none for the present default case).
The next four NBO output segments summarize the results of natural population analysis (NPA). The first segment is the main NAO table, as shown below:
NATURAL POPULATIONS: Natural atomic orbital occupancies
NAO Atom # lang Type(AO) Occupancy Energy
---------------------------------------------------------
1 C 1 s Cor( 1s) 1.99900 -11.04184
2 C 1 s Val( 2s) 1.09038 -0.28186
3 C 1 s Ryd( 3s) 0.00068 1.95506
4 C 1 px Val( 2p) 0.89085 -0.01645
5 C 1 px Ryd( 3p) 0.00137 0.93125
6 C 1 py Val( 2p) 1.21211 -0.07191
7 C 1 py Ryd( 3p) 0.00068 1.03027
8 C 1 pz Val( 2p) 1.24514 -0.08862
9 C 1 pz Ryd( 3p) 0.00057 1.01801
10 N 2 s Cor( 1s) 1.99953 -15.25950
11 N 2 s Val( 2s) 1.42608 -0.71700
12 N 2 s Ryd( 3s) 0.00016 2.75771
13 N 2 px Val( 2p) 1.28262 -0.18042
14 N 2 px Ryd( 3p) 0.00109 1.57018
15 N 2 py Val( 2p) 1.83295 -0.33858
16 N 2 py Ryd( 3p) 0.00190 1.48447
17 N 2 pz Val( 2p) 1.35214 -0.19175
18 N 2 pz Ryd( 3p) 0.00069 1.59492
19 H 3 s Val( 1s) 0.81453 0.13283
20 H 3 s Ryd( 2s) 0.00177 0.95067
21 H 4 s Val( 1s) 0.78192 0.15354
22 H 4 s Ryd( 2s) 0.00096 0.94521
23 H 5 s Val( 1s) 0.78192 0.15354
24 H 5 s Ryd( 2s) 0.00096 0.94521
25 H 6 s Val( 1s) 0.63879 0.20572
26 H 6 s Ryd( 2s) 0.00122 0.99883
27 H 7 s Val( 1s) 0.63879 0.20572
28 H 7 s Ryd( 2s) 0.00122 0.99883
For each of the 28 NAO functions, this table lists the atom to which the NAO is attached, the angular momentum type (s, px, etc.), the orbital type (whether core, valence, or Rydberg, with a conventional hydrogenic-type label), the orbital occupancy (number of electrons, or "natural population" of the orbital), and the orbital energy (in atomic units: 1 a.u. = 627.5 kcal/mol). Note that the occupancies of the Rydberg (Ryd) NAOs are typically much lower than those of the core (Cor) and valence (Val) NAOs of the natural minimum basis (NMB) set, reflecting the dominant role of the NMB orbitals in describing molecular properties.
The principal quantum numbers for the NAO labels (1s, 2s, 3s, etc.) are assigned on the basis of the energy if a Fock or Kohn-Sham matrix is available, or on the basis of occupancy otherwise. A message warning of a "population inversion" is printed if the occupancy and energy ordering do not coincide (of interest, but rarely of concern).
The next segment is an atomic summary showing the natural atomic charges (nuclear charge minus summed natural populations of NAOs on the atom) and total core, valence, and Rydberg populations on each atom:
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom # Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.44079 1.99900 4.43848 0.00331 6.44079
N 2 -0.89715 1.99953 5.89378 0.00384 7.89715
H 3 0.18370 0.00000 0.81453 0.00177 0.81630
H 4 0.21713 0.00000 0.78192 0.00096 0.78287
H 5 0.21713 0.00000 0.78192 0.00096 0.78287
H 6 0.35999 0.00000 0.63879 0.00122 0.64001
H 7 0.35999 0.00000 0.63879 0.00122 0.64001
=======================================================================
* Total * 0.00000 3.99853 13.98820 0.01328 18.00000
This table succinctly describes the molecular charge distribution in terms of NPA charges.
Next follows a summary of the NMB and NRB populations for the composite system, summed over atoms:
Natural Population
--------------------------------------------------------
Core 3.99853 ( 99.9632% of 4)
Valence 13.98820 ( 99.9157% of 14)
Natural Minimal Basis 17.98672 ( 99.9262% of 18)
Natural Rydberg Basis 0.01328 ( 0.0738% of 18)
--------------------------------------------------------
This reveals the high percentage contribution (typically, > 99%) of the NMB set to the molecular charge distribution.
Finally, the natural populations are summarized as an effective valence electron configuration ("natural electron configuration") for each atom:
Atom # Natural Electron Configuration ---------------------------------------------------------------------------- C 1 [core]2s( 1.09)2p( 3.35) N 2 [core]2s( 1.43)2p( 4.47) H 3 1s( 0.81) H 4 1s( 0.78) H 5 1s( 0.78) H 6 1s( 0.64) H 7 1s( 0.64)
Although the occupancies of the atomic orbitals are non-integer in the molecular environment, the effective atomic configurations can be related to idealized atomic states in "promoted" configurations.
The next segments of the output summarize the results of NBO analysis. The first segment reports on details of the search for an NBO natural Lewis structure:
NATURAL BOND ORBITAL ANALYSIS:
Occupancies Lewis Structure Low High
Occ. ------------------- ----------------- occ occ
Cycle Thresh. Lewis Non-Lewis CR BD 3C LP (L) (NL) Dev
=============================================================================
1(1) 1.90 17.95048 0.04952 2 6 0 1 0 0 0.02
-----------------------------------------------------------------------------
Structure accepted: No low occupancy Lewis orbitals
Normally, there is but one cycle of the NBO search. The table summarizes a variety of information for each cycle: the occupancy threshold for a "good" pair in the NBO search; the total populations of Lewis and non-Lewis NBOs; the number of core (CR), 2-center bond (BD), 3-center bond (3C), and lone pair (LP) NBOs in the natural Lewis structure; the number of low-occupancy Lewis (L) and high-occupancy (> 0.1e) non-Lewis (NL) orbitals; and the maximum deviation (Dev) of any formal bond order for the structure from a nominal estimate (NAO Wiberg bond index). The Lewis structure is accepted if all orbitals of the formal Lewis structure exceed the occupancy threshold (default = 1.90 electrons).
Next follows a more detailed breakdown of the Lewis and non-Lewis occupancies into core, valence, and Rydberg shell contributions:
WARNING: 1 low occupancy (<1.9990e) core orbital found on C 1 -------------------------------------------------------- Core 3.99853 ( 99.963% of 4) Valence Lewis 13.95195 ( 99.657% of 14) ================== ============================ Total Lewis 17.95048 ( 99.725% of 18) ----------------------------------------------------- Valence non-Lewis 0.03977 ( 0.221% of 18) Rydberg non-Lewis 0.00975 ( 0.054% of 18) ================== ============================ Total non-Lewis 0.04952 ( 0.275% of 18) --------------------------------------------------------
This shows the general quality of the natural Lewis structure description in terms of the percentage of the total electron density (e.g., in the above case, about 99.7%). The table also exhibits the relatively important role of the valence non-Lewis orbitals (i.e., the six valence antibonds, NBOs 23-28, listed below) relative to the extra-valence orbitals (the 13 Rydberg NBOs 10-22) in the slight departures from a localized Lewis structure model. The table also includes a warning about a carbon core orbital with slightly less than double occupancy.
Next follows the main listing of NBOs, displaying the form and occupancy of the complete set of orbitals that span the input AO space:
(Occupancy) Bond orbital/ Coefficients/ Hybrids
-------------------------------------------------------------------------------
1. (1.99858) BD ( 1) C 1- N 2
( 40.07%) 0.6330* C 1 s( 21.71%)p 3.61( 78.29%)
-0.0003 -0.4653 -0.0238 -0.8808 -0.0291
-0.0786 -0.0110 0.0000 0.0000
( 59.93%) 0.7742* N 2 s( 30.88%)p 2.24( 69.12%)
-0.0001 -0.5557 0.0011 0.8302 0.0004
0.0443 -0.0098 0.0000 0.0000
2. (1.99860) BD ( 1) C 1- H 3
( 59.71%) 0.7727* C 1 s( 25.78%)p 2.88( 74.22%)
-0.0002 -0.5077 0.0069 0.1928 0.0098
0.8396 -0.0046 0.0000 0.0000
( 40.29%) 0.6347* H 3 s(100.00%)
-1.0000 -0.0030
3. (1.99399) BD ( 1) C 1- H 4
( 61.02%) 0.7812* C 1 s( 26.28%)p 2.80( 73.72%)
0.0001 0.5127 -0.0038 -0.3046 -0.0015
0.3800 -0.0017 0.7070 -0.0103
( 38.98%) 0.6243* H 4 s(100.00%)
1.0000 0.0008
4. (1.99399) BD ( 1) C 1- H 5
( 61.02%) 0.7812* C 1 s( 26.28%)p 2.80( 73.72%)
0.0001 0.5127 -0.0038 -0.3046 -0.0015
0.3800 -0.0017 -0.7070 0.0103
( 38.98%) 0.6243* H 5 s(100.00%)
1.0000 0.0008
5. (1.99442) BD ( 1) N 2- H 6
( 68.12%) 0.8253* N 2 s( 25.62%)p 2.90( 74.38%)
0.0000 0.5062 0.0005 0.3571 0.0171
-0.3405 0.0069 -0.7070 -0.0093
( 31.88%) 0.5646* H 6 s(100.00%)
1.0000 0.0020
6. (1.99442) BD ( 1) N 2- H 7
( 68.12%) 0.8253* N 2 s( 25.62%)p 2.90( 74.38%)
0.0000 0.5062 0.0005 0.3571 0.0171
-0.3405 0.0069 0.7070 0.0093
( 31.88%) 0.5646* H 7 s(100.00%)
1.0000 0.0020
7. (1.99900) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
1.0000 -0.0003 0.0000 -0.0002 0.0000
0.0001 0.0000 0.0000 0.0000
8. (1.99953) CR ( 1) N 2 s(100.00%)p 0.00( 0.00%)
1.0000 -0.0001 0.0000 0.0001 0.0000
0.0000 0.0000 0.0000 0.0000
9. (1.97795) LP ( 1) N 2 s( 17.85%)p 4.60( 82.15%)
0.0000 0.4225 0.0002 0.2360 -0.0027
0.8749 -0.0162 0.0000 0.0000
10. (0.00105) RY*( 1) C 1 s( 1.57%)p62.84( 98.43%)
0.0000 -0.0095 0.1248 -0.0305 0.7302
-0.0046 0.6710 0.0000 0.0000
11. (0.00034) RY*( 2) C 1 s( 0.00%)p 1.00(100.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0146 0.9999
12. (0.00022) RY*( 3) C 1 s( 56.51%)p 0.77( 43.49%)
0.0000 -0.0023 0.7517 -0.0237 0.3710
-0.0094 -0.5447 0.0000 0.0000
13. (0.00002) RY*( 4) C 1 s( 41.87%)p 1.39( 58.13%)
14. (0.00116) RY*( 1) N 2 s( 1.50%)p65.53( 98.50%)
0.0000 -0.0062 0.1224 0.0063 0.0371
0.0197 0.9915 0.0000 0.0000
15. (0.00044) RY*( 2) N 2 s( 0.00%)p 1.00(100.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 -0.0132 0.9999
16. (0.00038) RY*( 3) N 2 s( 33.38%)p 2.00( 66.62%)
0.0000 0.0133 0.5776 0.0087 -0.8150
-0.0121 -0.0405 0.0000 0.0000
17. (0.00002) RY*( 4) N 2 s( 65.14%)p 0.54( 34.86%)
18. (0.00178) RY*( 1) H 3 s(100.00%)
-0.0030 1.0000
19. (0.00096) RY*( 1) H 4 s(100.00%)
-0.0008 1.0000
20. (0.00096) RY*( 1) H 5 s(100.00%)
-0.0008 1.0000
21. (0.00122) RY*( 1) H 6 s(100.00%)
-0.0020 1.0000
22. (0.00122) RY*( 1) H 7 s(100.00%)
-0.0020 1.0000
23. (0.00016) BD*( 1) C 1- N 2
( 59.93%) 0.7742* C 1 s( 21.71%)p 3.61( 78.29%)
-0.0003 -0.4653 -0.0238 -0.8808 -0.0291
-0.0786 -0.0110 0.0000 0.0000
( 40.07%) -0.6330* N 2 s( 30.88%)p 2.24( 69.12%)
-0.0001 -0.5557 0.0011 0.8302 0.0004
0.0443 -0.0098 0.0000 0.0000
24. (0.01569) BD*( 1) C 1- H 3
( 40.29%) 0.6347* C 1 s( 25.78%)p 2.88( 74.22%)
0.0002 0.5077 -0.0069 -0.1928 -0.0098
-0.8396 0.0046 0.0000 0.0000
( 59.71%) -0.7727* H 3 s(100.00%)
1.0000 0.0030
25. (0.00769) BD*( 1) C 1- H 4
( 38.98%) 0.6243* C 1 s( 26.28%)p 2.80( 73.72%)
-0.0001 -0.5127 0.0038 0.3046 0.0015
-0.3800 0.0017 -0.7070 0.0103
( 61.02%) -0.7812* H 4 s(100.00%)
-1.0000 -0.0008
26. (0.00769) BD*( 1) C 1- H 5
( 38.98%) 0.6243* C 1 s( 26.28%)p 2.80( 73.72%)
-0.0001 -0.5127 0.0038 0.3046 0.0015
-0.3800 0.0017 0.7070 -0.0103
( 61.02%) -0.7812* H 5 s(100.00%)
-1.0000 -0.0008
27. (0.00426) BD*( 1) N 2- H 6
( 31.88%) 0.5646* N 2 s( 25.62%)p 2.90( 74.38%)
0.0000 -0.5062 -0.0005 -0.3571 -0.0171
0.3405 -0.0069 0.7070 0.0093
( 68.12%) -0.8253* H 6 s(100.00%)
-1.0000 -0.0020
28. (0.00426) BD*( 1) N 2- H 7
( 31.88%) 0.5646* N 2 s( 25.62%)p 2.90( 74.38%)
0.0000 -0.5062 -0.0005 -0.3571 -0.0171
0.3405 -0.0069 -0.7070 -0.0093
( 68.12%) -0.8253* H 7 s(100.00%)
-1.0000 -0.0020
For each NBO (1-28), the first line of printout shows the occupancy (between 0 and 2 electrons) and unique label of the NBO. This label gives the type (BD for 2-center bond, CR for 1-center core pair, LP for 1-center valence lone pair, RY* for 1-center Rydberg, and BD* for 2-center antibond, the unstarred and starred labels corresponding to Lewis and non-Lewis NBOs, respectively), a serial number (1, 2,... if there is a single, double,... bond between the pair of atoms), and the atom(s) to which the NBO is affixed. The next lines summarize the natural atomic hybrids hA of which the NBO is composed, giving the percentage (cA-squared) of the NBO on each hybrid (in parentheses), the polarization coefficient cA, the atom label, and a hybrid label showing the sp-hybridization (percentage s-character, p-character, etc.) of each hA. Below each NHO label is the set of coefficients that specify how the NHO is written explicitly as a linear combination of NAOs on the atom. The order of NAO coefficients follows the numbering of the NAO tables.
In the CH3NH2 example, the NBO search finds the C-N bond (NBO 1), three C-H bonds (NBOs 2, 3, 4), two N-H bonds (NBOs 5, 6), N lone pair (NBO 9), and C and N core pairs (NBOs 7, 8) of the expected Lewis structure. NBOs 10-28 represent the residual non-Lewis NBOs of low occupancy. In this example, it is also interesting to note the slight asymmetry of the three sCH NBOs, and the slightly higher occupancy (0.016 vs. 0.008 electrons) in the sCH3 antibond (NBO 24) lying trans to the nitrogen lone pair.
The next segment of output summarizes the angular properties of the natural hybrid orbitals (NHOs):
NHO Directionality and "Bond Bending" (deviations from line of nuclear centers)
[Thresholds for printing: angular deviation > 1.0 degree]
hybrid p-character > 25.0%
orbital occupancy > 0.10e
Line of Centers Hybrid 1 Hybrid 2
--------------- ------------------- ------------------
NBO Theta Phi Theta Phi Dev Theta Phi Dev
===============================================================================
1. BD ( 1) C 1- N 2 90.0 5.4 -- -- -- 90.0 182.4 3.0
3. BD ( 1) C 1- H 4 35.3 130.7 34.9 129.0 1.0 -- -- --
4. BD ( 1) C 1- H 5 144.7 130.7 145.1 129.0 1.0 -- -- --
5. BD ( 1) N 2- H 6 144.7 310.7 145.0 318.3 4.4 -- -- --
6. BD ( 1) N 2- H 7 35.3 310.7 35.0 318.3 4.4 -- -- --
9. LP ( 1) N 2 -- -- 90.0 74.8 -- -- -- --
The "direction" of a hybrid is specified in terms of the polar (q) and azimuthal (f) angles (in the coordinate system of the calling program) of the vector describing its p-component. For more general spldm hybrids the hybrid direction is determined numerically to correspond to the maximum angular amplitude. The hybrid direction is then compared with the direction of the line of centers between the two nuclei to determine the bending of the bond, expressed as the deviation angle (Dev, in degrees) between these two directions. For example, in the CH3NH2 case shown above, the nitrogen NHO of the sCN bond (NBO 1) is bent away from the line of C-N centers by 3.0°, whereas the carbon NHO is approximately aligned with the C-N axis (within the 1.0° threshold for printing). The N-H bonds (NBOs 5, 6) are bent even further (by 4.4°). The information in this table is often useful in anticipating the direction of geometry changes resulting from geometry optimization (viz., likely reduced pyramidalization of the -NH2 group to relieve the ~4° nitrogen bond bending found in the tetrahedral Pople-Gordon geometry).
The next segment summarizes the second-order perturbative estimates of donor-acceptor (bond-antibond) interactions in the NBO basis:
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===============================================================================
within unit 1
2. BD ( 1) C 1- H 3 / 14. RY*( 1) N 2 0.84 2.18 0.038
3. BD ( 1) C 1- H 4 / 26. BD*( 1) C 1- H 5 0.52 1.39 0.024
3. BD ( 1) C 1- H 4 / 27. BD*( 1) N 2- H 6 3.03 1.37 0.057
4. BD ( 1) C 1- H 5 / 25. BD*( 1) C 1- H 4 0.52 1.39 0.024
4. BD ( 1) C 1- H 5 / 28. BD*( 1) N 2- H 7 3.03 1.37 0.057
5. BD ( 1) N 2- H 6 / 10. RY*( 1) C 1 0.56 1.78 0.028
5. BD ( 1) N 2- H 6 / 25. BD*( 1) C 1- H 4 2.85 1.51 0.059
6. BD ( 1) N 2- H 7 / 10. RY*( 1) C 1 0.56 1.78 0.028
6. BD ( 1) N 2- H 7 / 26. BD*( 1) C 1- H 5 2.85 1.51 0.059
7. CR ( 1) C 1 / 16. RY*( 3) N 2 0.61 13.11 0.080
7. CR ( 1) C 1 / 18. RY*( 1) H 3 1.40 11.99 0.116
7. CR ( 1) C 1 / 19. RY*( 1) H 4 1.55 11.99 0.122
7. CR ( 1) C 1 / 20. RY*( 1) H 5 1.55 11.99 0.122
8. CR ( 1) N 2 / 10. RY*( 1) C 1 1.51 16.23 0.140
8. CR ( 1) N 2 / 12. RY*( 3) C 1 0.84 16.77 0.106
8. CR ( 1) N 2 / 21. RY*( 1) H 6 0.61 16.26 0.089
8. CR ( 1) N 2 / 22. RY*( 1) H 7 0.61 16.26 0.089
9. LP ( 1) N 2 / 24. BD*( 1) C 1- H 3 8.13 1.13 0.086
9. LP ( 1) N 2 / 25. BD*( 1) C 1- H 4 1.46 1.14 0.037
9. LP ( 1) N 2 / 26. BD*( 1) C 1- H 5 1.46 1.14 0.037
This analysis is carried out by examining all possible interactions between "filled" (donor) Lewis-type NBOs and "empty" (acceptor) non-Lewis NBOs, and estimating their energetic importance by 2nd-order perturbation theory. Since these interactions lead to donation of occupancy from the localized NBOs of the idealized Lewis structure into the empty non-Lewis orbitals (and thus, to departures from the idealized Lewis structure description), they are referred to as "delocalization" corrections to the zeroth-order natural Lewis structure. For each donor NBO (i) and acceptor NBO (j), the stabilization energy E(2) associated with delocalization ("2e-stabilization") i ® j is estimated as
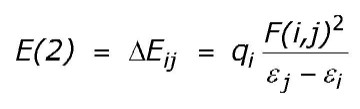
where qi is the donor orbital occupancy, ei, ej are diagonal elements (orbital energies) and F(i,j) is the off-diagonal NBO Fock matrix element. [In the example above, the nN ® sCH* interaction between the nitrogen lone pair (NBO 8) and the antiperiplanar C1-H3 antibond (NBO 24) is seen to give the strongest stabilization, 8.13 kcal/mol.] As the heading indicates, entries are included in this table only when the interaction energy exceeds a default threshold of 0.5 kcal/mol.
Next appears a condensed summary of the principal NBOs, showing the occupancy, orbital energy, and the qualitative pattern of delocalization interactions associated with each:
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
===============================================================================
Molecular unit 1 (CH5N)
1. BD ( 1) C 1- N 2 1.99858 -0.89908
2. BD ( 1) C 1- H 3 1.99860 -0.69181 14(v)
3. BD ( 1) C 1- H 4 1.99399 -0.68892 27(v),26(g)
4. BD ( 1) C 1- H 5 1.99399 -0.68892 28(v),25(g)
5. BD ( 1) N 2- H 6 1.99442 -0.80951 25(v),10(v)
6. BD ( 1) N 2- H 7 1.99442 -0.80951 26(v),10(v)
7. CR ( 1) C 1 1.99900 -11.04131 19(v),20(v),18(v),16(v)
8. CR ( 1) N 2 1.99953 -15.25927 10(v),12(v),21(v),22(v)
9. LP ( 1) N 2 1.97795 -0.44592 24(v),25(v),26(v)
10. RY*( 1) C 1 0.00105 0.97105
11. RY*( 2) C 1 0.00034 1.02120
12. RY*( 3) C 1 0.00022 1.51414
13. RY*( 4) C 1 0.00002 1.42223
14. RY*( 1) N 2 0.00116 1.48790
15. RY*( 2) N 2 0.00044 1.59323
16. RY*( 3) N 2 0.00038 2.06475
17. RY*( 4) N 2 0.00002 2.25932
18. RY*( 1) H 3 0.00178 0.94860
19. RY*( 1) H 4 0.00096 0.94464
20. RY*( 1) H 5 0.00096 0.94464
21. RY*( 1) H 6 0.00122 0.99735
22. RY*( 1) H 7 0.00122 0.99735
23. BD*( 1) C 1- N 2 0.00016 0.57000
24. BD*( 1) C 1- H 3 0.01569 0.68735
25. BD*( 1) C 1- H 4 0.00769 0.69640
26. BD*( 1) C 1- H 5 0.00769 0.69640
27. BD*( 1) N 2- H 6 0.00426 0.68086
28. BD*( 1) N 2- H 7 0.00426 0.68086
-------------------------------
Total Lewis 17.95048 ( 99.7249%)
Valence non-Lewis 0.03977 ( 0.2209%)
Rydberg non-Lewis 0.00975 ( 0.0542%)
-------------------------------
Total unit 1 18.00000 (100.0000%)
Charge unit 1 0.00000
This table allows one to quickly identify the principal delocalizing acceptor orbitals associated with each donor NBO, and their topological relationship to this NBO, i.e., whether attached to the same atom (geminal, "g"), to an adjacent bonded atom (vicinal, "v"), or to a more remote ("r") site. These acceptor NBOs will generally correspond to the principal "delocalization tails" of the NLMO associated with the parent donor NBO. [For example, in the table above, the nitrogen lone pair (NBO 9) is seen to be the lowest-occupancy (1.978 electrons) and highest-energy (-0.446 a.u.) Lewis NBO, and to be primarily delocalized into antibonds 24, 25, 26 (the vicinal sCH NBOs). The summary at the bottom of the table shows that the Lewis NBOs 1-9 describe about 99.7% of the total electron density, with the remaining non-Lewis density found primarily in the valence-shell antibonds (particularly, NBO 24).]